MCS 275 Spring 2022 Project 1¶
- Course instructor: Emily Dumas
Instructions¶
Deadline is 6pm CDT on Friday 4 February 2022.¶
Note that the hour of the deadline is not the same as for homework assignments.
Collaboration policy and academic honesty¶
This project must be completed individually. Seeking or giving aid on this assignment is prohibited; doing so constitutes academic misconduct which can have serious consequences. The only resources you are allowed to consult are the ones listed below. If you are unsure about whether something is allowed, ask. The course syllabus contains more information about the course and university policies regarding academic honesty.
Topic¶
In this project you will read a substantial amount of existing object-oriented code in order to understand how to use it. You will then add a few new subclasses that interact with the existing code.
The classes in this project form an object-oriented bacteria simulation.
Resources you are allowed to consult¶
- Documents and videos posted to the course web page
- Any of the optional textbooks listed on the course web page
Ask if you are unsure whether a resource falls under one of these categories, or if you think I missed an essential resource.
What to do if you are stuck¶
Contact the instructor or your TA by email, in office hours, or on discord.
Get the starter pack¶
The starter pack is available here:
It is a ZIP archive, i.e. a group of files bundled together and compressed into a single file. The first thing you need to do is to extract the starter pack into a directory where you will do your work. Contact the instructor or TA if you need assistance extracting a ZIP file.
What's in the starter pack¶
It contains A FILE YOU EDIT AND SUBMIT AS YOUR PROJECT:
bacteria.py- ContainsBacteriumclass; you need to add three more classes as described below.
And also:
Two supporting files that I provide, and which you don't edit at all:
environments.py- A module containing classes for different types of environments bacteria can live in.plane.py- The 2D vector and point module we developed in Lectures 4 and 5 (which are used extensively in this project).
This sample program that shows how you can test the bacterial simulation:
textsimulation.py
Concept overview¶
Reading this section will allow you to understand the goals and key concepts of the project. To start writing code you'll also need the technical documentation in the next section>
Bacteria¶
One of the object-oriented programming examples developed in lecture involves a series of simulated robots that move in the plane according to various rules. This project is similar in spirit, but involves bacteria moving and reproducing in an environment. Unlike the robot simulation example from lecture, the bacteria in this simulation need to consume resources from their environment, and so there is a separate class that manages these resources.
The environment¶
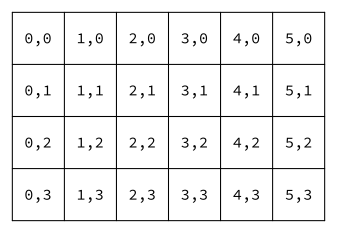
The environment on which bacteria grow is modeled by a rectangular grid where each location is labeled by a pair of integers (x,y). The value of x indicates the column, and the value of y indicates the row, each starting at 0. The coordinates of grid squares are shown below for a 6x4 grid, i.e. a grid with 6 columns and 4 rows.
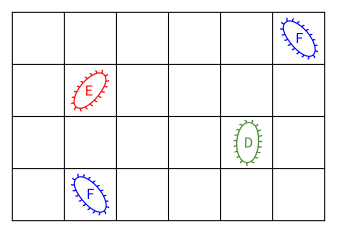
A grid square can be empty, or it can be occupied by a bacterium. The environment keeps track of a list of the bacteria that currently live on it. The picture below depicts a environment with a few bacteria (of three types, labeled "D", "E", and "F"). Each of these bacteria would have a corresponding object that manages its behavior (and you will write the classes that do this).
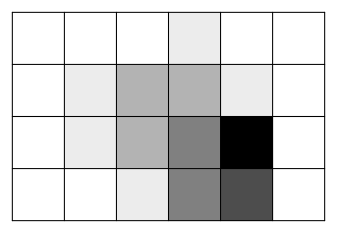
Each grid square also contains a certain quantity of nutrients. The quantity is an integer between 0 and 8 (inclusive). The nutrient quantity is an abstraction of all the resources a bacterium absorbs from its environment. The behavior of a bacterium in the environment will depend on the available nutrients. In the image below, an example distribution of nutrients is shown by coloring squares in shades of gray, with white meaning no nutrients at all, and progressively darker shades of gray indicating cells with larger quantities of nutrients.
environments.py¶
The file environments.py from the starter pack contains a class Environment that represents an environment of the type described above. It manages the list of bacteria that are present, and the quantity of nutrients in each grid square. Initially, every square has the same quantity of nutrients. An environment is created as
Environment(6,4) # 6 columns, 4 rows, initially all have 8 nutrientsor
Environment(6,4,initial_nutrients=3) # customize initial nutrient quantityAn environment with a uniform distribution of nutrients is not so interesting, so this file also contains some subclasses that do more interesting things:
HillEnvironment- an environment where the middle is nutrient-rich, while the edges have a low nutrient densityGradientEnvironment- an environment with more nutrients near the bottom (larger y coordinate), less near the top (smaller y coordinate).
bacteria.py¶
This file contains just one class, Bacterium. It represents a bacterium that lives in an environment. The environment must already exist, and is given as an argument to the constructor of Bacterium.
The Bacterium class takes an initial position, adds itself to the environment, and then just sits there doing nothing. The point of this project is to make subclasses that represent bacteria with more interesting behavior.
Specifically, you are going to build these subclasses:
Eater- A bacterium that consumes nutrients at its location until they are exhausted, then moves to a neighbhoring grid square that contains nutrients. If no such neighboring square exists, it dies.Floater- A bacterium that always tries to decrease its y coordinate by moving up one row. If it can't do that, it consumes one unit of nutrients. If neither of those things is possible, it dies.Divider- A bacterium that can't move, but which can reproduce if conditions are right (plenty of nutrients and available space). If it can't reproduce, it eats. If it can't do either of those things, it dies.
These short descriptions don't provide enough information to write the classes. See the section Specification of the classes you must write below for more information.
Documentation of classes provided for you¶
This section contains technical documentation for all the code in the starter pack.
Do not edit any of the classes described here. Reading the source code is a great idea, though!
Classes in environments.py¶
class Environment¶
Superclass: None
Purpose: Represent a bacterial growth environment that starts with a constant nutrient amount in each grid square. Keep track of all the bacteria living in the environment. Advance the bacterial simulation one time step on request.
Attributes meant to be available to users of the class:
width: The width of the grid, in columns, as given to the constructorheight: The height of the grid, in rows, as given to the constructororganisms: A list of bacteria that are currently part of the simulation, in the order they were added. Dead bacteria are removed from this list after each step of the simulation.
Methods:
__init__(self,width,height,initial_nutrients=8): Make a new environment of sizewidthcolumns andheightrows. Each grid square initially contains a quantity of nutrients given byinitial_nutrients.is_valid_position(self,p): ReturnsTrueif the Point2 objectpa valid grid square, i.e. if0 <= p.x < self.widthand0 <= p.y < self.height.is_available(self,p): ReturnsTrueif the Point2 objectpis a valid grid square that is currently unoccupied.get_nutrients(self,p): Returns the amount of nutrients at grid squarep(a Point2 object), an integer between 0 and 8 inclusive.set_nutrients(self,p,k): Sets the amount of nutrients atpto the valuek.consume_nutrients(self,p,k): Subtractskfrom the amount of nutrients atp, as long as the result is not negative. If a negative number would result,ValueErroris raised instead.add(self,o): Add the bacteriumoto this environment. (The constructor ofBacteriumcalls this to add itself.)update(self): Advance the simulation by one time step, by calling the.update()method of each element ofself.organisms(in the order they appear in that list) and then removing dead bacteria from the list.
Note: All of these methods except add and update are meant to be used by subclasses of Bacterium to plan and implement their actions (e.g. checking for available nutrients, moving, ...).
class GradientEnvironment¶
Superclass: Environment
Purpose: Represents an environment with a nutrient gradient - lots at the bottom, little at the top. Same methods as Environment, differs only in the initial quantities of nutrients setup by the constructor.
class HillEnvironment¶
Superclass: Environment
Purpose: Represents an environment with a nutrient gradient - lots at the middle, few at the edges. Same methods as Environment, differs only in the initial quantities of nutrients setup by the constructor.
Classes in bacteria.py¶
class Bacterium¶
Superclass: None
Purpose: Represents a bacterium that sits in one place (in a given environment) forever. Meant to be subclassed to give interesting behaviors.
Attributes meant to be available to users of the class:
env- The environment this bacterium lives inposition- A Point2 object giving the bacterium's locationalive- A boolean indicating whether this bacterium is alive (don't modify directly; usedie()method)
Methods:
__init__(self,env,position): Make a new bacterium at positionpositionand add to environmentenv.can_move(self,v): Is it allowable for this bacterium to move by Vector2v? (That is, to addvto its currentpositionattribute?) Check with the environment to determine the answer and return a boolean. Allowable means the position is valid and unoccupied.move(self,v): Change the bacterium's position by adding Vector2vto itspositionnutrients_available(self,v=Vector2(0,0)): Return quantity of nutrients available at the bacterium's position, or ifvis specified, return the quantity available atself.position + v. (Used to check current or nearby nutrient amounts.)consume(self,k): Consumeknutrients at the current position (raises ValueError if not enough nutrients are available.die(self): Make this bacterium die.update(self): Advance the simulation one time step. Does nothing in this class, but is meant to be redefined in subclasses to implement their unique behaviors.
Specification of the classes you must write¶
Everything you write goes in bacteria.py. You need to create three new subclasses of Bacterium that do the things described below. Don't edit Bacterium itself, though; just add new subclasses.
class Eater¶
Superclass: Bacterium
Purpose: Represents a bacterium that eats until it runs out of nutrients, moving only when necessary.
Methods to be defined in this subclass:
update(self): The action depends on whether the current location contains any nutrients:- If at least one unit of nutrients is available: Consume one unit.
- If no nutrients are available: Look at the neighboring grid cells, i.e. the ones obtained by adding either
Vector2(1,0),Vector2(-1,0),Vector2(0,1), orVector2(0,-1)to the position. Consider them in that order, and move to the first one that is found to contain at least one unit of nutrients. If none of the neighbor cells have any nutrients, die.
Do not add any other methods. Do not copy code from Bacterium; use inheritance instead.
class Floater¶
Superclass: Bacterium
Purpose: Represents a bacterium that always tries to move upward.
Methods to be defined in this subclass:
update(self): The action depends on whether it is possible to move byVector2(0,-1)("up"):- If it is possible: Move by
Vector2(0,-1). - If it is not possible: Eat one unit of nutrients. If none are available, die.
- If it is possible: Move by
Do not add any other methods. Do not copy code from Bacterium; use inheritance instead.
class Divider¶
Superclass: Bacterium
Purpose: Represents a bacterium that cannot move, but which can reproduce if conditions are just right.
Methods to be defined in this subclass:
update(self): The action depends on whether the conditions are right for reproduction. The required conditions for reproduction are:- The current location has at least 3 units of nutrients, and
- One of the neighboring locations (current position plus
Vector2(1,0),Vector2(-1,0),Vector2(0,1), orVector2(0,-1)) is empty. (These are tried in the order shown here, and the first empty one found is remembered.)
If the reproduction conditions are satisfied, consume 3 units of nutrients and create a new instance of
Dividerthat lives in the same environment and which is located at the first empty neighbor location identified above. If the reproduction conditions are not satisfied (e.g. too few nutrients or no available space), consume 1 unit of nutrients if possible. If no nutrients are available, die.
Do not add any other methods. Do not copy code from Bacterium; use inheritance instead.
Sample usage¶
Here is a simple program that makes a few bacteria and runs a simulation (with no visualization):
from plane import Vector2, Point2
from bacteria import Eater, Floater, Divider # Note: you write these classes!
from environments import HillEnvironment
env = HillEnvironment(10,10) # 10x10 grid with nutrients near the middle
Eater(env,Point2(7,7)) # Make an Eater bacterium and add to the environment
Divider(env,Point2(6,6)) # Make a Divider bacterium and add to the environment
Floater(env,Point2(3,8)) # Make a Floater bacterium and add to the environment
t=0
while env.organisms: # While any are still alive...
env.update()
t += 1
print("At time {} there are {} bacteria alive".format(t,len(env.organisms)))
While it's a little hard to tell what's going on from the table above, it is clear that more bacteria are created for a while but then they start to die off.
The situation becomes clearer if you make a visualization of the simulation. The animated GIF below represents the same simulation as in the sample program above, with shades of gray showing nutrients, eaters in red, floaters in blue, and dividers in green. The floaters and dividers die out quickly (after an initial period where the dividers flourish), and one lonely eater is left for the last 48 steps.

Demonstration program¶
The program textsimulation.py included in the starter pack has the ability to run a simulation and display it using simple text graphics. You'll want to edit the program to add some new bacteria, because as included in the starter pack, it only creates a few Bacterium objects (which don't move or do anything). As you build new subclasses of Bacterium, I recommend trying them out by adding them to the simulation. You'll find some commented-out lines that give suggested bacteria to add when available.
You can just run
python3 textsimulation.pyto see a simulation like this:
Bacterial simulation in 40x8 environment
.,,,::; D DDD#@@F@@@@@###lllii;;::,,,.
.,,,::;;iiFDD##@@@@@@@@@@###llii;;::,,,.
.,,:::;;iill###@@DDD@@@@@###llii;;:::,,.
,,, E:;;iill###DDDD DDDDDllii;;:::,,,
,,,:::;;iill###@DDDDDD DDD##llii;;:::,,,
.,,:::;;iill###@@@DDDDDD@###llii;;:::,,.
.,,,::;;iill###@@@@@DD@@@###llii;;::,,,.
,,,::;;iilll###@@@@@@@@###lllii;;::,,,.
7 steps, 44 organisms alive
ENTER for next time stepThe characters .,:;il#@ represent increasing amounts of nutrients, and the letters B, E, F, D represent Bacterium, Eater, Floater, and Divider objects, respectively.
Optionally, if your terminal and font support unicode bar graph characters, you can select fancier text-base visualization by running
python3 textsimulation.py --fancywhich will look like
Bacterial simulation in 40x8 environment
▁▂▂▂▃▃▄ D DDD▇██F█████▇▇▇▆▆▆▅▅▄▄▃▃▂▂▂▁
▁▂▂▂▃▃▄▄▅▅FDD▇▇██████████▇▇▇▆▆▅▅▄▄▃▃▂▂▂▁
▁▂▂▃▃▃▄▄▅▅▆▆▇▇▇██DDD█████▇▇▇▆▆▅▅▄▄▃▃▃▂▂▁
▂▂▂ E▃▄▄▅▅▆▆▇▇▇DDDD DDDDD▆▆▅▅▄▄▃▃▃▂▂▂
▂▂▂▃▃▃▄▄▅▅▆▆▇▇▇█DDDDDD DDD▇▇▆▆▅▅▄▄▃▃▃▂▂▂
▁▂▂▃▃▃▄▄▅▅▆▆▇▇▇███DDDDDD█▇▇▇▆▆▅▅▄▄▃▃▃▂▂▁
▁▂▂▂▃▃▄▄▅▅▆▆▇▇▇█████DD███▇▇▇▆▆▅▅▄▄▃▃▂▂▂▁
▂▂▂▃▃▄▄▅▅▆▆▆▇▇▇████████▇▇▇▆▆▆▅▅▄▄▃▃▂▂▂▁
7 steps, 44 organisms alive
ENTER for next time stepif the required characters are available.
IMPORTANT RULES YOUR CODE MUST FOLLOW¶
Your project must follow the 7 rules in the MCS 275 coding standards document. In addition:
- The methods you add to subclasses of
Bacteriumshould only call other methods of theBacteriumclass and build-in Python functions. It should not be necessary to call methods ofEnvironmentdirectly, as theBacteriumclass provides everything you need to interact with the environment (e.g.move(),nutrients_available(),consume(), anddie()).
- Only submit
bacteria.py. The autograder will supply unmodified versions ofenvironment.pyandplane.pyto use with it when testing your project.
- The only things you add to
bacteria.pyshould be class definitions. Do not add any code outside of class definitions (no functions that are not methods, no test code, etc.)
- Make proper use of inheritance by calling methods of
Bacteriumwhen it is necessary to do things like move, check whether movement is possible, check nutrient amounts, consume nutrients, or die.
How your project will be graded¶
Autograder: 50 points¶
The autograder tests your program and grades it based on its behavior. The following tests will be run:
- Was a file called
bacteria.pysubmitted? (5 points) - Does the Python interpreter accept the contents of
bacteria.pyas valid Python code? (5 points) - Does
bacteria.pycontain docstrings for all classes, functions, and methods? (5 points) - Can
bacteria.pybe imported without raising an exception, in a directory containing theenvironments.pyandplane.pyfiles from the starter pack? (5 points) - Does
bacteria.pycontain classes namedEater,Floater, andDividerthat are all subclasses ofBacterium? (6 points) - Functional tests: Do the classes
Eater,Floater, andDividerbehave as specified when placed in environments and simulated? (several test cases totaling 24 points)
Manual review: 10 points¶
I will review your code and look for adherence to the coding standards and other rules given above, and I will examine your method of solving the problem. If I see that your program does not do what was requested in this document, but the error was not detected in the automated testing, a deduction may be given at this point. The scores assigned by the autograder will not be changed during manual review unless I discover some kind of intentional wrongdoing (such as an attempt to circumvent or reverse-engineer the autograder's operation).
Final word: It is really important to test locally¶
As you write your project, test it locally on your own computer or the lab computer you use for your work. Every time you create a new bacterium class, add an instance of it to textsimulation.py and try it out. It is much harder to debug a broken program based solely on reports you get from the autograder compared to working with your local Python interpreter.
Revision history¶
- 2022-01-24 Initial publication